Bacteria in uranium wastes and their interactions with uranium
Bacteria are the most dominating organisms in the biosphere
They occur in water and soil not only on the surface but also in the deep terrestrial subsurface. They occupy niches such as hot springs, Antarctic ice, alkaline lakes, acidic liquors, salt lakes, deserts etc., where the life of plants, animals and insects is impossible. Bacteria are distributed also in environments which are highly contaminated with radionuclides and heavy metals. It was demonstrated that the levels of radioactivity and radionuclides even in highly contaminated soil or water samples do not have a significant negative effect on the bacteria indigenous for these environments. Moreover, many bacterial species are involved in:
Biotransformation of toxic metals:
Some bacterial species can oxidise particular metals or metal sulphides transforming them into a soluble form. Other species, in contrast, are involved in reduction and precipitation of some heavy metals and radionuclides. Many bacteria are able also to bioaccumulate U, Co, Cs, Sr, Mo, Pb, Hg, Cu, Mn, Al, Cd, Ga, Ba and other toxic metals to varying degrees and to transport them outside of the polluted environments. A large number of highly polluted with heavy metals and radionuclides environments in Europe are stemming from the uranium mining and uranium production. These hazardous wastes are one of the most severe environmental problems at present. In many European countries (for example, Spain, Germany, United Kingdom, France, Italy, Czech, Romania, Bulgaria, Russia, Ukraine, Poland) uranium mining was or still is intensively performed. At the uranium waste piles in all these countries there are soils, water, and sediments which are contaminated with uranium and other hazardous metals such as Se, Mo, As, Cd, Cr, Hg, Pb, Cu, Ni, Cs, Sr, Zn, etc. At the places where the extraction of uranium from the ores was performed (at the so named "mill tailings") significant amounts of Th, Ra, and Po might also be present.
How to remediate the environments polluted with heavy metals?
Conventional chemical and physical methods for removing dissolved heavy metals include chemical precipitation, chemical oxidation and reduction, ion exchange, and filtration. These processes are very expensive. They may also be ineffective when applied to decontaminate uranium wastes in natural conditions, in particular because such wastes have an extremely complex composition. The extremely complicated biotic and abiotic environmental factors are, however, not a limiting factor for the activity of the natural bacterial communities in these environments, because they are well adapted to them.
The prospective for developing reasonably cheap and effective bacteria-based techniques for the removal of metals from polluted effluents, waste water, sediments, and soils requires extensive analyses of the structure and of the activity of the natural bacterial communities in the contaminated wastes. It is necessary to clear who are those bacteria which are adapted "to work" effectively there. On the second hand, it is very important to study the biological mechanisms of microbial accumulation and reduction of heavy metals by which those bacteria are doing the "decontamination work". And last but not least, it is of greatest interest to find out which are the genes involved in the expression and in the controlling of such functions. The latter is of highest interest, because it will provide scientists with a possibility not only to learn more about the mechanisms of metal uptake and reduction by bacteria and to use them more effectively. It might also provide a possibility to improve these functions by manipulations of the genes, which are responsible for them and, possibly, to construct bacterial strains which will be useful for a development of biotechnologies for bioremediation of the uranium wastes.
The goal of our studies in the Institute of Radiochemistry, FZD is:
To find out how to use the potential of bacteria in treating heavy metals and radionuclides in the hazardous uranium waste piles.
For this reason we are analysing diversity and activity in several uranium wastes in Saxony - the "Haberland Halde" near Johanngeorgenstadt, and the mill tailings near Steinsee Deponie 1, Gitersee/Coschüth and Schlema/Alberoda. Applying several modern molecular biological approaches, such as Ribosomal Intergenic Spacer Amplification (RISA) and 16S ribosomal DNA (rDNA) Amplification, Pulsed-field Gel Electrophoresis (PFGE), Random Amplified Polymorphic DNA (RAPD), Repetive Primer Amplified Polymorphic DNA (rep-APD), as well as Time-resolved Laser Fluorescence Spectroscopy (TRLFS), we have found that extremely high bacterial diversity occurs in the samples investigated. In particular, the presence of several dominant 16S rDNA groups related to the genera Thiobacillus, Bacillus, Desulfovibrio and Pseudomonas was demonstrated.
Thiobacillus ferrooxidans
One of these 16S rDNA groups was affiliated to the species Thiobacillus ferrooxidans. The strains of this species are chemolithoautotrophic, they can oxidise U4+ and Fe2+ as well as also many different metal sulphides from the natural minerals. The latter is known as bioleaching or solubilization of metals. These bacteria can also accumulate uranium in acidic liquors (pH 1,5). Interestingly, we have found three different 16S rDNA types corresponding to the species T. ferrooxidans. These types possess slightly different 16S rDNA patterns. Strains of T. ferrooxidans possessing two of the mentioned three 16S rDNA types were recovered from two soil samples polluted in different extend with heavy metals. The two groups of isolates have different genomic organization than the known reference strains.
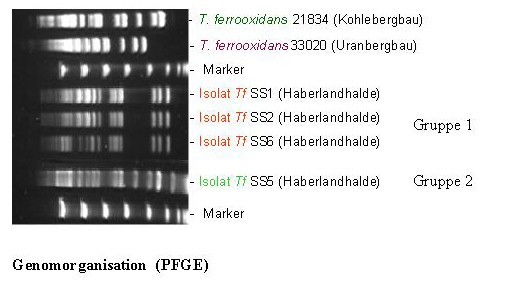
In addition, the members of the group recovered from the more polluted sample are tolerant to higher concentrations of uranyl ions which are lethal for the isolates of the second group. The expression of at least three genes of the U-tolerant strains is influenced by the presence of uranyl ions in the nutrient medium. The capability of the U-mine isolates to interact with U(VI) was studied, and it was found that they accumulate significantly higher amounts of U(VI) in comparison to several reference T. ferrooxidans strains recovered from other environments. Using time-resolved laser fluorescence spectroscopy it was shown that the complexes build by one of the U-waste isolates with U(VI) are much stronger than those build by the reference strains.
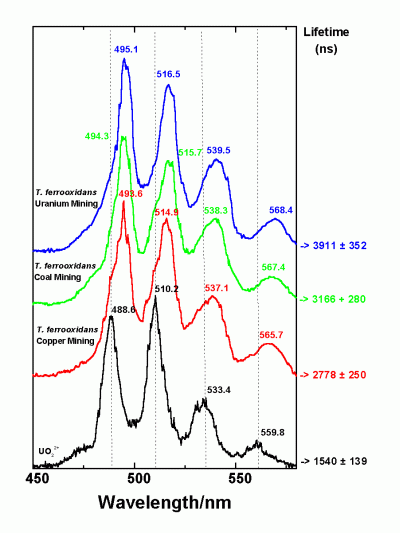
Time-resolved laser fluorescence spectroscopy of Uranium onto T. ferrooxidans
This is the first demonstration of microdiversity in closely related natural isolates of T. ferrooxidans. We suggest that the microdiversity observed reflects the genetic adaptation of the strains studied to the different heavy metal concentrations in their natural environments.
Bacillus
The second uranium waste bacterial group belongs to the genus Bacillus. We have classified three of the Bacillus waste isolates, as B. cereus, B. megaterium and B. sphaericus, and we have demonstrated that they are able to selectively accumulate different heavy metals from drain waters of the "Haberland Halde".
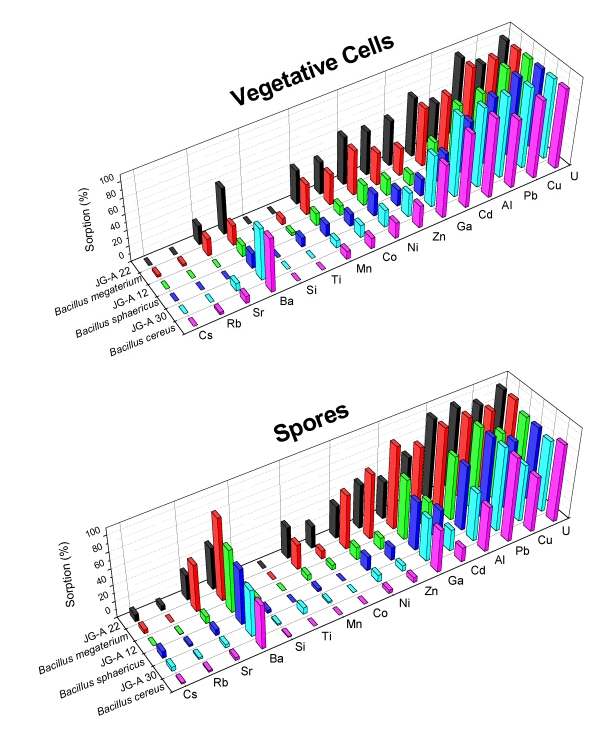
Selective bioaccumulation of heavy metals in groundwater
We have found that the Bacillus strains are binding significantly higher amounts of uranium at pH 4.8 in comparison to the above mentioned T. ferrooxidans strains. However, the complexes built between the Bacillus isolates and uranium are much weaker than those built by the T. ferrooxidans strains. The latter was confirmed using TRLFS as well.
S-Layer
One of the uranium waste isolates, JG-A12, possesses a novel kind of S-Layer protein.
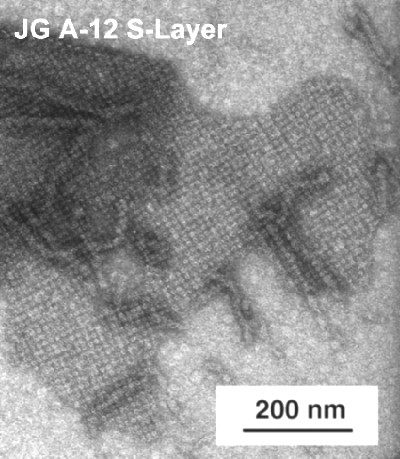
TEM micrograph of negatively stained S-Layer from the Bacillus sphaericus isolate JG A-12, recovered from a uranium mining waste pile.
S-Layers are porous crystalline protein membranes 5-15 nm thick, which completely cover the cell surface and can provide microorganisms with a selective advantage by functioning as protective coats, molecular sieves, molecule and ion traps, etc. They possess a great potential for biotechnology, medicine and nanotechnology.
Desulfovibrio
The third group of uranium waste isolates belongs to the genus Desulfovibrio. These sulphate-reducing bacteria are able also to reduce U(VI) to U(IV). Up to date we have analysed two uranium waste Desulfovibrio isolates which were recovered in Umweltforschungszentrum Leipzig. These isolates are closely related to the species Desulfovibrio vulgaris subspecies (oxamicus).
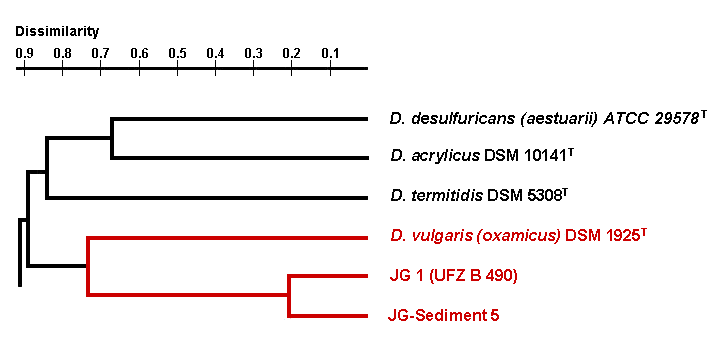
Dendrogram of the phylogenetical relationships of Desulfovibrio strains
In addition their genomic organization differs significantly from those of the type strain of this species. In addition, the analysis of the kinetic of the uranium reduction by one of the Desulfovibrio isolates has shown that the waste isolate is able to reduce significantly faster larger amounts of U(VI) than the reference Desulfovibrio strain. In contrast to the reference strain, the waste isolate reduced uranium independently on the pH range of the medium. To our knowledge, no other bacterium of the genus Desulfovibrio is able to reduce and precipitate U(VI) from solutions so rapidly and effectively as this isolate.
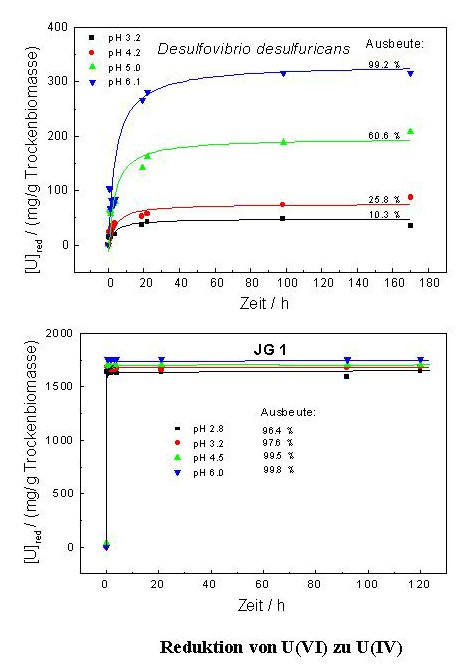
The natural uranium waste bacterial isolates analysed in our laboratory might be of great importance for the development of bioremediation procedures for the environments polluted with uranium and other heavy metals, because they are well adapted to their extremely complex geological, chemical and biological conditions.

